(cover image from ThisisEngineering RAEng)
Let’s face it: software is easier to write than maintain. This is why we, as software engineers, prefer to just “rip it out and start over” instead of trying to understand what another developer (or our past self) was thinking. We seem to have collectively forgotten that “programs must be written for people to read, and only incidentally for machines to execute”. You know it’s true — we’ve all had to painstakingly trace through a casserole of spaghetti code and thin, old-world-style abstractions digging for the meat of the program only to find nothing but a mess at the bottom of our plates.
It’s easy to yell “WTF” and blame the previous dev, but the truth is often more complicated. We can’t see the future, so it’s impossible to understand how requirements, technology, or business goals will grow when we design a net-new system. As a result, systems can become unreadable as their scope increases along with the business’s dependency on them. This is a bit of a paradox: older, harder to maintain systems often provide the most value. They are hard to work on because they’ve grown with the company, and scary to work on because breaking it could be a catastrophe.
Here’s where I’m calling you out: if you like hard, rewarding problems… try it. Take the oldest system you have and make it maintainable. You know the one I’m talking about — the one no one will “own”. That one the other departments depend on but engineers hate. The one you had to patch Log4Shell on first. Do it. I dare you.I recently had such an opportunity to update a decade old machine learning system at Bazaarvoice. On the surface, it didn’t sound exciting: this thing didn’t even have neural networks! Who cares! We ll… it mattered. This system processes nearly every user-generated product review received by Bazaarvoice — nearly 9 million per month — and does so with 90 million inference calls to machine learning models. Yup — 90 million inferences! It’s a huge scale, and I couldn’t wait to dive in.
In this post, I’ll share how modernizing this system through a re-architecture, instead of a re-write, allowed us to make it scalable and cost-effective without having to rip out all of the code and start over. The resulting system is serverless, containerized, and maintainable while reducing our hosting costs by nearly 80%. This post is a companion piece to a talk I recently presented at AWS Data Summit for Software Companies on generating value from data by leveraging our best practices to ensure success in machine learning projects. This post discusses the technical aspects in more detail, but you can watch the high-level talk linked at the end.
Something Old
First, let’s take a look at what we’re dealing with here. The legacy system my team was updating moderates user-generated content for all of Bazaarvoice. Specifically, it determines if each piece of content is appropriate for our client’s websites.
This sounds straightforward — eliminate obvious infractions such as hate speech, foul language, or solicitations — but in practice, it’s much more nuanced. Each client has unique requirements for what they consider appropriate. Beer brands, for example, would expect discussions of alcohol, but a children’s brand may not. We capture these client-specific options when we onboard new clients, and our Client Services team encodes them into a management database.
For some added complexity, we also sample a subset of our content to be moderated by human moderators. This allows us to continuously measure the performance of our models and discover opportunities for building more models.
The full architecture of our legacy system is shown below:
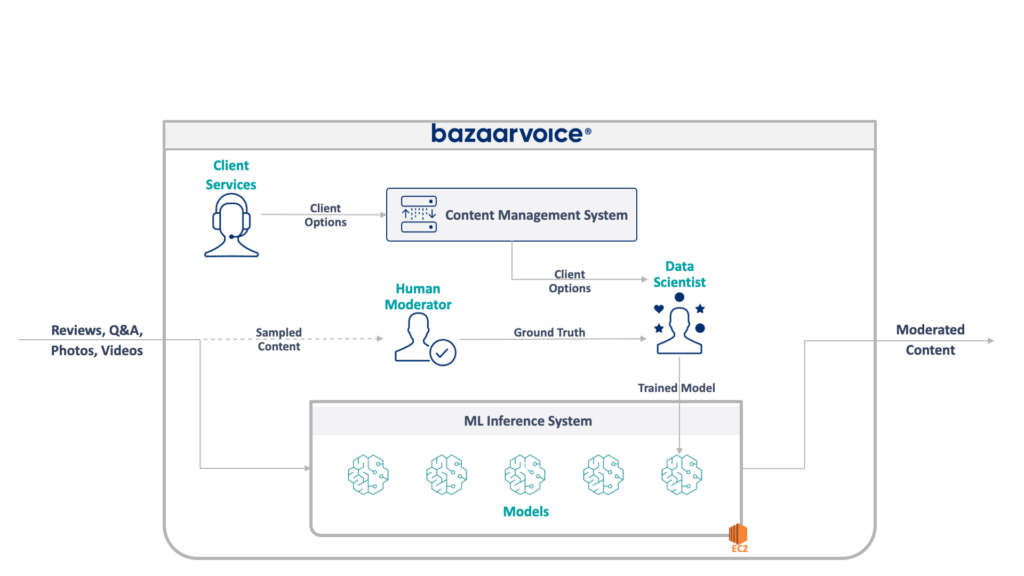
This system has some serious drawbacks. Specifically — all of the models are hosted on a single EC2 instance. This wasn’t due to bad engineering — just the inability of the original programmers to foresee the scale desired by the company. No one thought that it would grow as much as it did. In addition, the system suffered from developer rejection: it was written in Scala, which few engineers understood. Thus, it was often overlooked for improvement since no one wanted to touch it.
As a result, the system continued to grow in a keep-the-lights-on manner. Once we got around to re-architecting it, it was running on a single x1e.8xlarge instance. This thing had nearly a terabyte of ram and costs about $5,000/month (unreserved) to operate. Don’t worry, though, we just launched a second one for redundancy and a third for QA 🙃.
This system was costly to run and was at a high risk of failure (a single bad model can take down the whole service). Furthermore, the code base had not been actively developed and was thus significantly out of date with modern data science packages and did not follow our standard practices for services written in Scala.
Something New
When redesigning this system we had a clear goal: make it scalable. Reducing operating costs was a secondary goal, as was easing model and code management.
The new design we came up with is illustrated below:
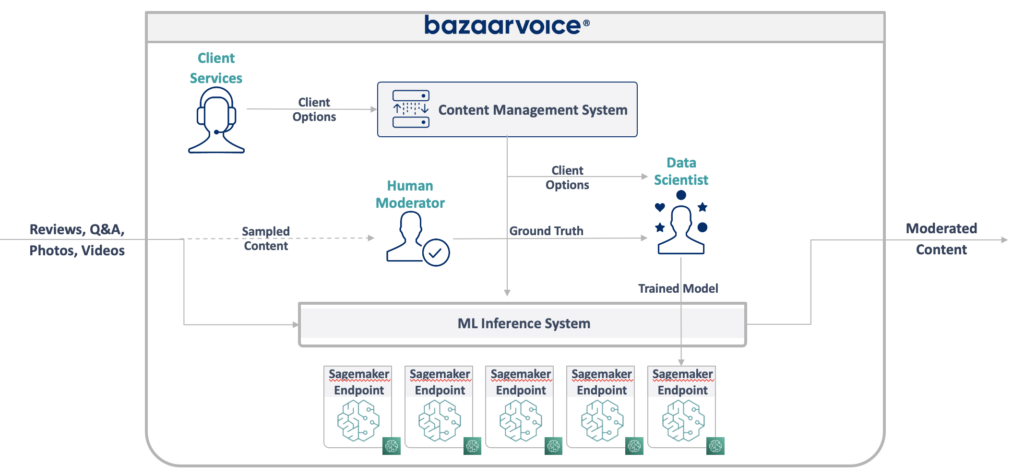
Our approach to solving all of this was to put each machine learning model on an isolated SageMaker Serverless endpoint. Like AWS Lambda functions, serverless endpoints turn off when not in use — saving us runtime costs for infrequently used models. They also can scale out quickly in response to increases in traffic.
In addition, we exposed the client options to a single microservice that routes content to the appropriate models. This was the bulk of the new code we had to write: a small API that was easy to maintain and let our data scientists more easily update and deploy new models.
This approach has the following benefits:
- Decreased the time to value by over 6x. Specifically, routing traffic to existing models is instantaneous, and deploying new models can be done in under 5 minutes instead of 30.
- Scale without limit – we currently have 400 models but plan to scale to thousands to continue to increase the amount of content we can automatically moderate.
- Saw a cost reduction of 82% moving off EC2 as the functions turn off when not in use, and we’re not paying for top-tier machines that are underutilized.
Simply designing an ideal architecture, however, isn’t the really interesting hard part of rebuilding a legacy system — you have to migrate to it.
Something Borrowed
Our first challenge in migration was figuring out how the heck to migrate a Java WEKA model to run on SageMaker, let alone SageMaker Serverless.
Fortunately, SageMaker deploys models in Docker containers, so at least we could freeze the Java and dependency versions to match our old code. This would help ensure the models hosted in the new system returned the same results as the legacy one.

To make the container compatible with SageMaker, all you need to do is implement a few specific HTTP endpoints:
POST /invocation— accept input, perform inference, and return results.GET /ping— returns 200 if the JVM server is healthy
(We chose to ignore all of the cruft around BYO multimodel containers and the SageMaker inference toolkit.)
A few quick abstractions around com.sun.net.httpserver.HttpServer and we were ready to go.
And you know what? This was actually pretty fun. Toying with Docker containers and forcing something 10 years old into SageMaker Serverless had a bit of a tinkering vibe to it. It was pretty exciting when we got it working — especially when we got the legacy code to build it in our new sbt stack instead of maven. The new sbt stack made it easy to work on, and containerization ensured we could get proper behavior while running in the SageMaker environment.
Something Blue
So we have the models in containers and can deploy them to SageMaker — almost done, right? Not quite.

The hard lesson about migrating to a new architecture is that you must build three times your actual system just to support migration.
In addition to the new system, we had to build:
- A data capture pipeline in the old system to record inputs and outputs from the model. We used these to confirm that the new system would return the same results.
- A data processing pipeline in the new system to compute results and compare them to the data from the old system. This involved a large amount of measurement with Datadog and needed to offer the ability to replay data when we found discrepancies.
- A full model deployment system to avoid impacting the users of the old system (which would simply upload models to S3). We knew we wanted to move them to an API eventually, but for the initial release, we needed to do so seamlessly.
All of this was throw-away code we knew we could toss once we finished migrating all of the users, but we still had to build it and ensure that the outputs of the new system matched the old.
Expect this upfront.
While building the migration tools and systems certainly took more than 60% of our engineering time on this project, it too was a fun experience. Unit testing became more like data science experiments: we wrote whole suites to ensure that our output matched exactly. It was a different way of thinking that made the work just that much more fun. A step outside our normal boxes, if you will.
So… Just Try It
Next time you’re tempted to rebuild a system from code up, I’d like to encourage you to try migrating the architecture instead of the code. You’ll find interesting and rewarding technical challenges and will likely enjoy it much more than debugging unexpected edge cases of your new code.
The talk I gave is a bit more high-level and goes into the MLOps side of things. Check it out here:

